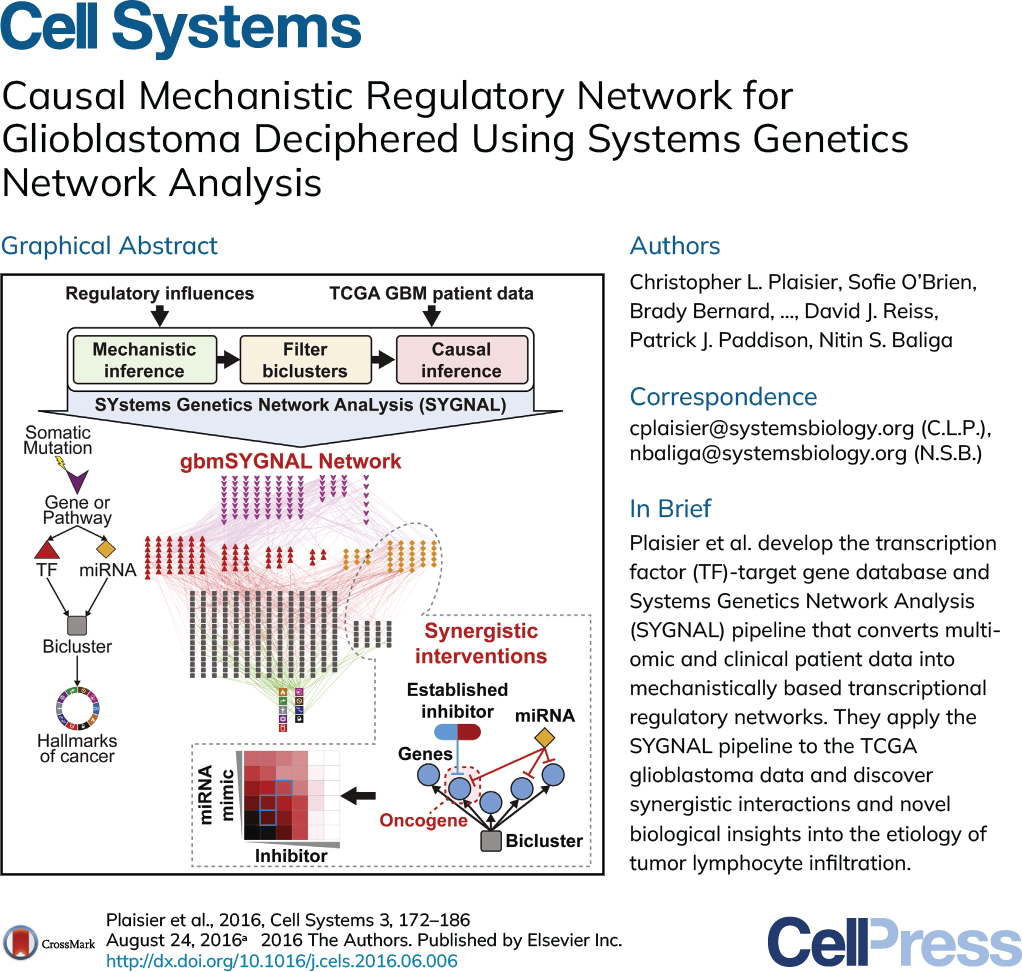
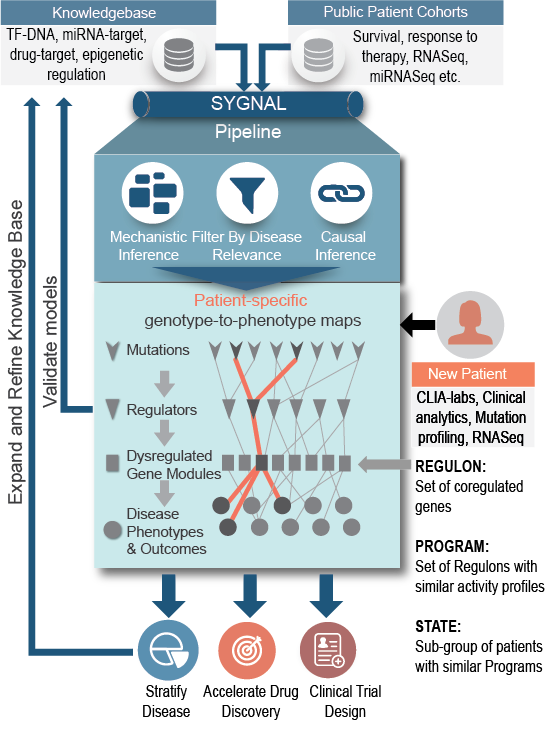
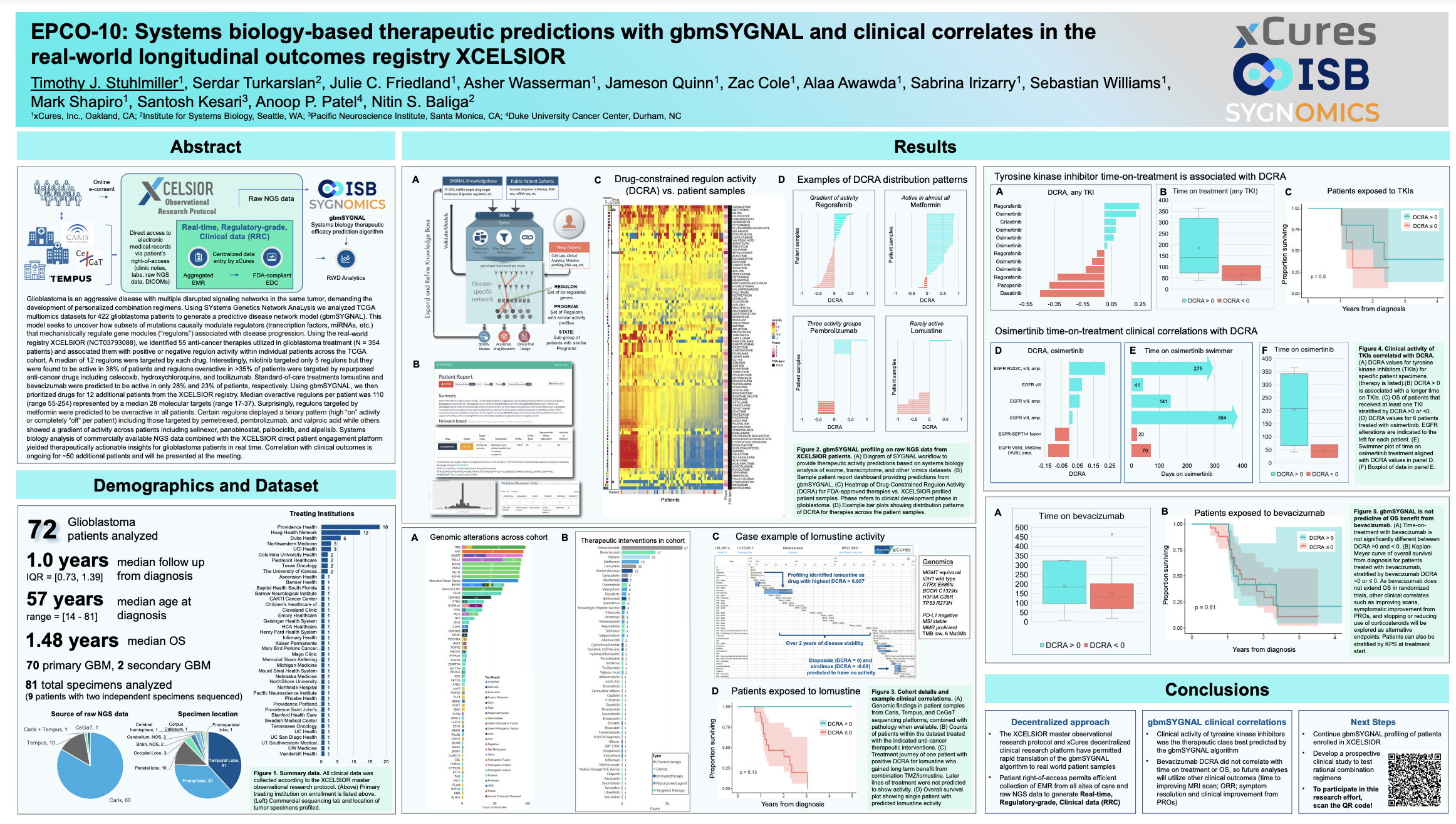
Our Approach
SYstems Genetics Network AnaLysis (SYGNAL)
Cancer is a complex disease harboring a mixture of bystander or passenger mutations (inactive) and driver mutations (active) that act through downstream effects on gene expression and network activity. Due to this complex mixture of active and inactive mutations, biomarkers based on correlates in mutations and simple gene expression patterns are seldom indicative of the causal and mechanistic drivers of disease etiology, progression, and severity. This is why just <10% of mutations in any given cancer are actionable, making mutation-based therapy recommendations inaccurate and less frequently useful. Hence the current paradigm of personalized precision oncology builds on a limited view of matching patients to therapies based on the presence of specific mutations in their tumor cells. This approach limits the universe of actionable insights by ignoring all of the biology that is downstream of DNA mutations. Therefore, novel approaches that incorporate multiple types of information (DNA, transcriptome, epigenome) into a cohesive framework are urgently needed to improve the success rates of precision medicine approaches.
Using the SYGNAL technology developed at the Institute for Systems Biology (ISB), Sygnomics uses statistical inference and machine learning to mine multiomics cancer datasets to delineate how subsets of mutations act causally and mechanistically to alter transcriptional programs governing disease phenotypes. Most importantly, by means of causal and mechanistic connections from mutations, through regulators, to co-regulated gene modules (aka regulons), the SYGNAL model provides meaningful and human interpretable links between many important but disconnected features that have been associated with cancer biology in the literature. Thus, a SYGNAL model can take both exome-seq and RNA-seq data of an individual patient to 1. Determine if a mutation is a bystander or driver by examining downstream consequences, and 2. Identify actionable therapeutic strategies that are independent of mutations and are based on gene expression and network activity.
Sygnomics ranks clinically actionable choices in the context of an individual patient
Patient-Specific Disease-Network Map
Network quantization will uncover patient-specific disease-network map to enable risk prediction and therapy prioritization.
By definition, it is not possible to infer statistically significant associations across mutations and gene expression patterns from an individual patient’s tumor biopsy. We have developed a novel concept to uncover the disease-perturbed network map (i.e., programs) within an individual patient by leveraging the SYGNAL model constructed from a patient cohort.
We have demonstrated that this approach is more robust and powerful than stratifying patients based on their cytogenetic markers, somatic mutations, or relative gene expression patterns. Because the programs are rooted in a causal and mechanistic disease network map for a given patient, we can use the activity of these programs for individualized risk assessment and also to prioritize therapies for that patient —this is why the SYGNAL network-based approach is innovative and transformational for personalized precision medicine.
Strong Systems Biology research is the foundation of the SYGNOMICS Knowledgebase.
GBM Network Portal
GBM Network Portal was developed to provide easy access to the gbmSYGNAL network by allowing searching of somatically mutated genes, regulators, or genes in biclusters.